Investigations in Human Sweat & Breath for New Biomarker Discovery
Vishnu Shankar, Zhenpeng Zhou
Non-invasive methods, compared to traditional medical diagnostics, are pain-free, cheaper, and more efficient, which is useful for early-detection, treatment, and illuminating important patient-level biochemical data. For example, investigations of human breath [1] revealed decreased levels of isoprene concentration for patients with chronic heart failure, which led to new connections between repair mechanisms in cholesterol biosynthesis and heart failure. In the same way, it has been suggested that human sweat [2], consisting of fatty acids, glycerides, and other lipid-based compounds, can aid novel insights in the metabolism for patients, which is measurably altered under disease states. Therefore, the objective of this study is to investigate human sweat and breath to find novel disease biomarkers.
Our current methodology, summarized in Figure 1, for extracting biomarkers from human sweat consists of three steps sample collection, collecting spectra through desorption electrospray ionization mass spectrometry (DESI-MS), and analyzing peaks through statistical learning methods.
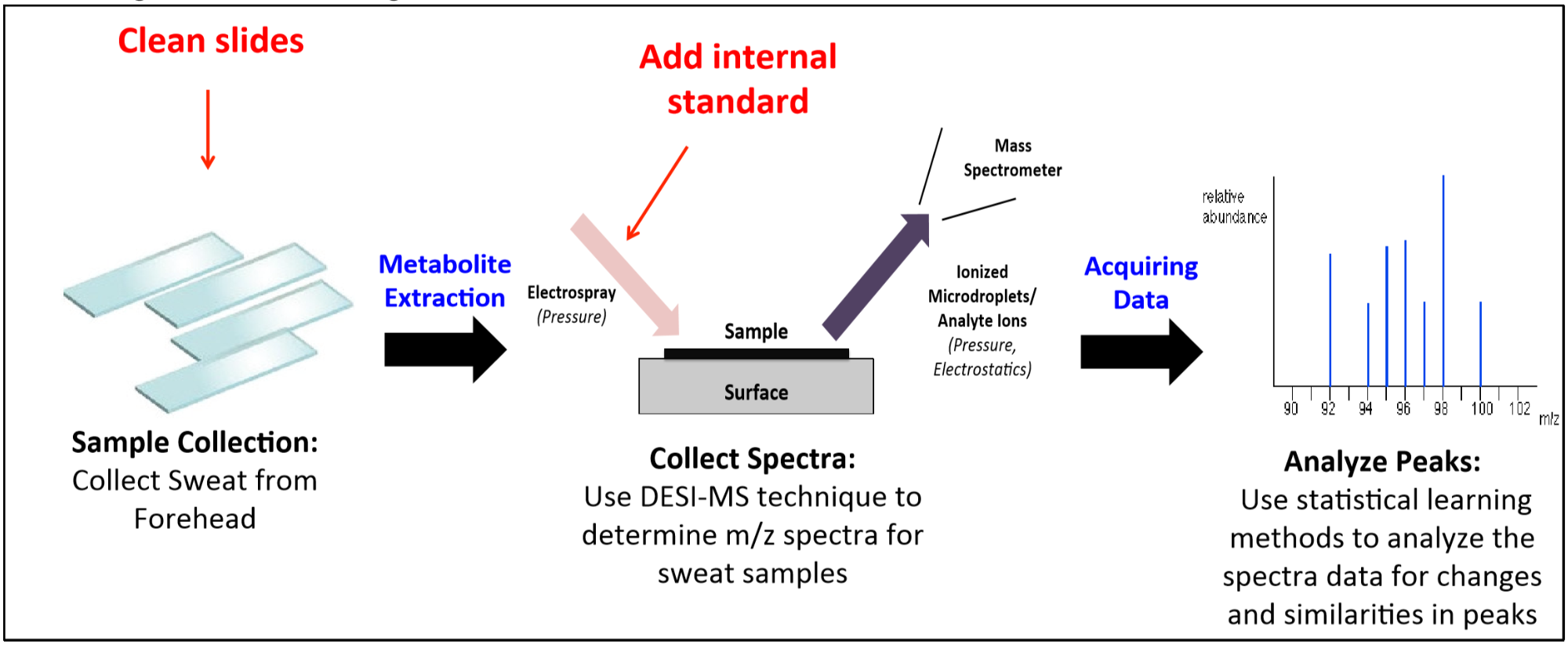
Figure 1. Summarized methodology to extract new biomarkers from human sweat
DESI-MS, a recently developed method, allows samples to be examined in ambient conditions, with minimal sample preparation. In DESI, a spray of charged droplets impacts a solid sample on a substrate and creates a thin liquid film. An additional splash of the incoming droplets creates microdroplets, which are then drawn into the mass spectrometer. This method offers lot of potential for rapid and routine real-time analysis along with sensitivity to detect small metabolites, fatty-acids, lipids, and heavy metabolite species. DESI-MS, in conjunction with statistical learning methods, can be leveraged to identify unique differences between different mass spectra for the purpose of biomarker discovery. For instance, Zhou and Zare (3) used DESI-MS in conjunction with gradient-boosting tree methods to classify individual gender, ethnicity, and age with over 80% accuracy in 194 sweat samples. Further, their analysis was able to use statistical methods and tandem mass-spectrometry to identify specific fatty acids (e.g. dialkylglycerol(16:1|10:0)) that were important in the classification.
In collaboration with the Stanford School of Medicine and Veterans Affairs Hospital in Palo Alto, we are currently investigating whether a similar methodology can be applied to find unique lipids and metabolites from patient sweat and breath that are relevant to disease-specific biochemical pathways. We hope this kind of analysis can spur new biomarker discovery, related to individual patient risk for metabolic conditions.
1. Miekisch, W., Schubert, J. K., & Noeldge-schomburg, G. F. E. (2004). Diagnostic potential of breath analysis — focus on volatile organic compounds. Clinica Chimica Acta, 347, 25–39. http://doi.org/10.1016/j.cccn.2004.04.023
2. Andres, P.; Zugaj, D.; Laffet, G.; Schoot, B. M.; Martel, P. Personal Identification Based on Sebum Composition. WO2011154259 A1, December 15, 2011.
3. Zhou, Z., & Zare, R. N. (2017). Personal Information from Latent Fingerprints Using Desorption Electrospray Ionization Mass Spectrometry and Machine Learning. ACS Analytical Chemistry, 89, 1369–1372. http://doi.org/10.1021/acs.analchem.6b04498.
Sample’Collec
